Single Genotype Circuit
Persistent Identifier
Use this permanent link to cite or share this Morpheus model:
Introduction
This model combines simple Notch signaling with the cellular Potts model (CPM), which drives cell sorting as a result of differential adhesion. Model details are contained in the reference Mulberry and Keshet, 2020.
In this experiment, we consider a mixture of cells of a single genotype. These cells can become ‘activated’ through lateral inhibition and eventually self-organize into a stable two-layer structure.
Description
We model a mixture of cells which all have both sender and receiver capabilities:

The full Delta-Notch dynamics are given by the following system of differential equations:
$$\begin{align} \frac{\mathrm dN}{\mathrm dt} &= N_0 \left(1 + \frac{I^p}{I_0^p+I^p}\right) - \kappa_tND_\text{ext}-\gamma N \\ \frac{\mathrm dI}{\mathrm dt} &= \kappa_tND_\text{ext}- \gamma_I I\\ \frac{\mathrm dE}{\mathrm dt} &= E_0 \frac{I^p}{I_0^p+I^p} - \gamma E\\ \frac{\mathrm dD}{\mathrm dt} &= D_0 - \kappa_tN_\text{ext}D - \gamma D \\ \end{align}$$
where $N$ denotes the Notch level in each cell, $D$ denotes the Delta level in each cell, $I$ is the Notch intracellular domain, and finally $E$ is the cell’s E-cad production. $N_\text{ext}$ and $D_\text{ext}$ are the average Notch and Delta levels of the cell’s immediate neighbourhood, respectively.
Results
Cells (initially all red) aggregate into a single cluster quickly and are gradually induced by neighbors to differentiate into more adhesive (green) E-cadherin expressing cells, forming differentiated structures. The structures are dynamic, with cells continuously changing type. Since the process of cell differentiation continues (i.e. cell states are not fixed), the structure is imperfect; however, once a cell differentiates, it is quickly sorted into its respective layer.
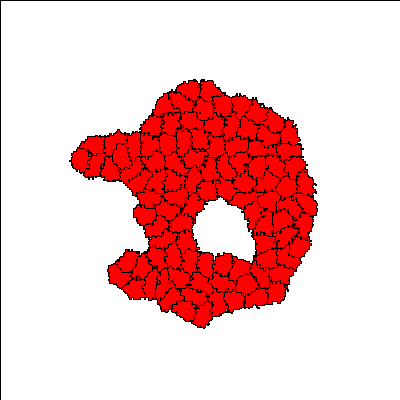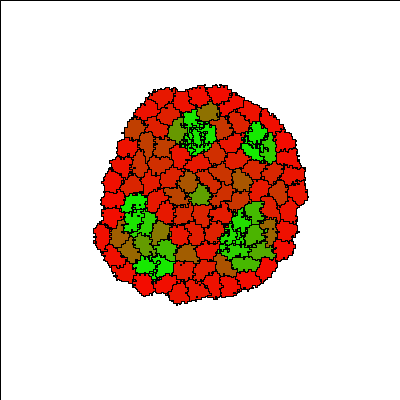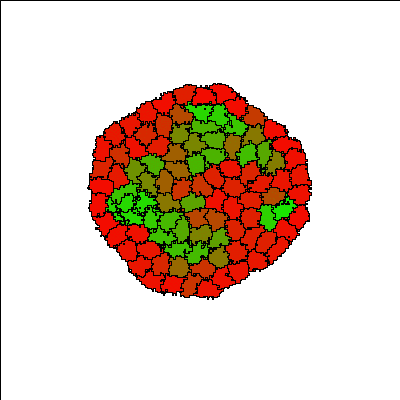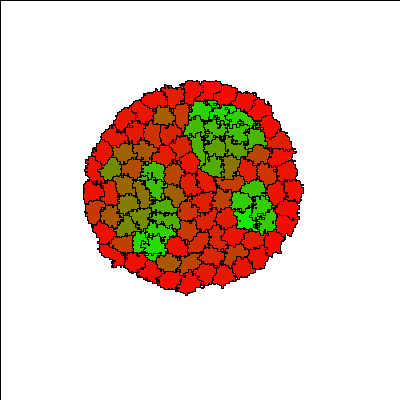
Reference
This model is described in the peer-reviewed publication:
N. Mulberry, L. Edelstein-Keshet: Self-organized Multicellular Structures from Simple Cell Signaling: A Computational Model. Phys. Biol. 17: 066003, 2020.
Our computational model was inspired by the experimental work of Toda et al., 2018. The Notch signaling model was based on that of Boareto et al., 2015.
Model
SingleGenotypeCircuit.xml
XML Preview
<MorpheusModel version="4">
<Description>
<Title>Single Genotype to Two Layer Circuit</Title>
<Details>
Formation of two-layer structure starting from a single genotype.
All cells initially the same, but they specialize into receivers and senders.
Reference:
The model is decribed in "Self-organized multicellular structures from simple cell signaling: a computational model",
N.Mulberry and L.Edelstein-Keshet, Physical Biology 2020, https://doi.org/10.1088/1478-3975/abb2dc
</Details>
</Description>
<Global>
<Variable symbol="N" value="0"/>
<Variable symbol="D" value="0"/>
<Variable symbol="Nn" value="0"/>
<Variable symbol="Dn" value="0"/>
<Variable symbol="I" value="0"/>
<Variable symbol="E" value="0"/>
<Variable symbol="Ad" value="0"/>
</Global>
<Space>
<SpaceSymbol symbol="l"/>
<Lattice class="square">
<Size symbol="size" value="300, 300, 0"/>
<BoundaryConditions>
<Condition boundary="x" type="noflux"/>
<Condition boundary="y" type="noflux"/>
</BoundaryConditions>
<Neighborhood>
<Order>1</Order>
</Neighborhood>
</Lattice>
</Space>
<Time>
<StartTime value="0"/>
<StopTime value="2e2"/>
<TimeSymbol symbol="time"/>
</Time>
<CellTypes>
<CellType class="biological" name="A">
<VolumeConstraint target="200" strength="1"/>
<Property symbol="N" value="1e3" name="notch"/>
<Property symbol="Nn" value="0.0" name="notch_Neighbours"/>
<Property symbol="D" value="3e3" name="delta"/>
<Property symbol="Dn" value="0.0" name="delta_Neighbours"/>
<Property symbol="I" value="0.0" name="NICD"/>
<Property symbol="E" value="0.0" name="Ecad"/>
<Property symbol="Ad" value="0.0" name="adhesive"/>
<NeighborhoodReporter>
<Input scaling="cell" value="N"/>
<Output symbol-ref="Nn" mapping="average"/>
</NeighborhoodReporter>
<NeighborhoodReporter>
<Input scaling="cell" value="D"/>
<Output symbol-ref="Dn" mapping="average"/>
</NeighborhoodReporter>
<System solver="runge-kutta" time-scaling="1" time-step="0.1">
<Constant symbol="Kc" value="5e-4"/>
<Constant symbol="Kt" value="5e-5"/>
<Constant symbol="N0" value="500"/>
<Constant symbol="D0" value="1000"/>
<Constant symbol="I0" value="200"/>
<Constant symbol="gammaI" value="0.5"/>
<Constant symbol="gamma" value="0.1"/>
<Constant symbol="p" value="2"/>
<DiffEqn symbol-ref="N">
<Expression>N0*(1+I^p/(I0^p+I^p)) - Kc*N*D - Kt*N*Dn - gamma*N</Expression>
</DiffEqn>
<DiffEqn symbol-ref="I">
<Expression>Kt*N*Dn - gammaI*I </Expression>
</DiffEqn>
<DiffEqn symbol-ref="D">
<Expression> D0*(I0^p/(I0^p+I^p)) - Kc*N*D - Kt*D*Nn - gamma*D</Expression>
</DiffEqn>
<DiffEqn symbol-ref="E">
<Expression> I^p/(I0^p+I^p) - gamma*E </Expression>
</DiffEqn>
<Constant symbol="Null" value="0.0" name="null"/>
</System>
</CellType>
<CellType class="medium" name="medium"/>
</CellTypes>
<CPM>
<Interaction>
<Contact type1="A" type2="medium" value="22.0"/>
<Contact type1="A" type2="A" value="16.0">
<HomophilicAdhesion strength="-1.6" adhesive="E"/>
</Contact>
</Interaction>
<MonteCarloSampler stepper="edgelist">
<MCSDuration value="0.001"/>
<Neighborhood>
<Order>2</Order>
</Neighborhood>
<MetropolisKinetics temperature="10"/>
</MonteCarloSampler>
<ShapeSurface scaling="norm">
<Neighborhood>
<Order>6</Order>
</Neighborhood>
</ShapeSurface>
</CPM>
<CellPopulations>
<Population size="0" type="A">
<InitCircle mode="random" number-of-cells="100">
<Dimensions radius="size.x/3" center="size.x/2, size.y/2, 0"/>
</InitCircle>
</Population>
</CellPopulations>
<Analysis>
<Logger time-step="1">
<Input>
<Symbol symbol-ref="N"/>
<Symbol symbol-ref="D"/>
<Symbol symbol-ref="I"/>
<Symbol symbol-ref="E"/>
<Symbol symbol-ref="Ad"/>
</Input>
<Output>
<TextOutput/>
</Output>
</Logger>
<Gnuplotter time-step="1" decorate="false">
<Terminal size="800 400 0" persist="true" name="png"/>
<Plot>
<Cells value="E" min="0.0" max="10.0">
<ColorMap>
<Color value="8.0" color="green"/>
<Color value="1.0" color="red"/>
</ColorMap>
</Cells>
</Plot>
</Gnuplotter>
<ClusteringTracker time-step="1.0" celltype="A"/>
</Analysis>
</MorpheusModel>
Downloads
Files associated with this model: