Neural Stem Cell Redivisions in Adult Zebrafish Telencephalon
Introduction
Whether individual stem cell divisions are coordinated or random is important for tissue morphogenesis, homeostasis and tumour growth. Lupperger et al. have analyzed the spatio-temporal division patterns of adult neural stem cells (NSC) in the zebrafish telencephalon. Imaging of over 80 000 NSCs in 36 brain hemispheres and comparison to model simulations with Morpheus revealed that redivisions (rapid cell cycle reentry by some newly born NSCs) can explain the observed aggregated division pattern.
Description
- Units in the model are
- Parameter values were fitted by Lupperger et al. to in vivo labelling experiments.
p_div=p_rediv=Global). The differentiation process is set to - Initial condition:
- The published model file was originally developed with Morpheus version 2.0.1 and is shared by the authors.
- The new model file provided below has been updated (solver naming, added reference and links to description, moved parameters
p_rediv,init_volumefromCellTypedefinition toGlobalto ease future modifications of their values) such that it works with Morpheus version 2.2.3 and later and reproduces the original results.
Results
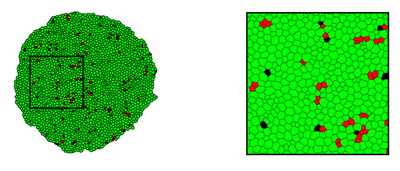
The model simulates the growth of the zebrafish telencephalon in a 2D cross section and visualizes NSCs in green, S-phase NSCs in red and progenitor cells in black as was presented in the published Figure S4G,H (shown below).
 ([Figure S4G,H](https://doi.org/10.1371/journal.pbio.3000708.s004)). [*CC BY 4.0*](https://creativecommons.org/licenses/by/4.0/)](https://d33wubrfki0l68.cloudfront.net/60ac7754d9abf08f03528a2fd5e6d24da9ecb808/a7327/media/model/m2984/original_fig.s4gh_huafd3ddec7998dcaa25fa2cacd61b0de1_272888_6bd5d2fcf73829f14bf0bf9482402b09.png)
Running the model for 150 000 Monte Carlo steps (1 500 hours of brain development) yields 2 726 cells and reproduces the aggregated division pattern.
plot-1*.png and plot-2*.pdf output files.
The spatiotemporal dynamics from 20 000-150 000 Monte Carlo steps is shown in the following Video. The left panel shows each cell clone in a unique color and the right panel shows the cell states color-coded as in the above Figures.
Reference
This model is the original used in the publication, up to technical updates:
V. Lupperger, C. Marr, P. Chapouton: Reoccurring neural stem cell divisions in the adult zebrafish telencephalon are sufficient for the emergence of aggregated spatiotemporal patterns. PLoS Biology 18 (12): e3000708, 2020.
Model
model.xml
XML Preview
<?xml version='1.0' encoding='UTF-8'?>
<MorpheusModel version="4">
<Description>
<Title>Lupperger2020</Title>
<Details>Full title: Neural stem cell (NSC) redivisions in adult zebrafish telencephalon
Date: 20.07.2021
Authors: V. Lupperger, C. Marr, P. Chapouton
Curators: V. Lupperger, L. Brusch
Software: Morpheus (open-source), download from https://morpheus.gitlab.io
ModelID: https://identifiers.org/morpheus/M2984
Reference: This model is described in the peer-reviewed publication
V. Lupperger, C. Marr, P. Chapouton. Reoccurring neural stem cell divisions in the adult zebrafish telencephalon are sufficient for the emergence of aggregated spatiotemporal patterns. PLoS Biology 18 (12): e3000708, 2020.
https://doi.org/10.1371/journal.pbio.3000708
</Details>
</Description>
<Global>
<Constant symbol="p_rediv" value="0.38"/>
<Variable symbol="c" value="0.0"/>
<Variable symbol="cellCycleTime" value="1e20"/>
<Variable symbol="time_of_birth" value="0.0"/>
<Variable symbol="p" value="2"/>
<Variable symbol="progenitor" value="0"/>
<Constant symbol="init_volume" value="50"/>
<Variable symbol="volume" value="init_volume"/>
<Variable symbol="minCellSize" value="25"/>
<Variable symbol="maxCellSize" value="250"/>
<Variable symbol="clone" value="0"/>
<Variable symbol="cc" value="0"/>
</Global>
<Space>
<Lattice class="hexagonal">
<Size symbol="size" value="1500, 1500, 0"/>
<BoundaryConditions>
<Condition type="periodic" boundary="x"/>
<Condition type="periodic" boundary="y"/>
</BoundaryConditions>
<Neighborhood>
<Order>3</Order>
<!-- <Disabled>
<Distance>1</Distance>
</Disabled>
-->
</Neighborhood>
<Annotation>spatial unit of one lattice step is \mathrm{\mu m}</Annotation>
</Lattice>
<SpaceSymbol symbol="space"/>
</Space>
<Time>
<StartTime value="0"/>
<StopTime symbol="stoptime" value="1.5e5">
<Annotation>time unit is \mathrm{0.01 hours}</Annotation>
</StopTime>
<TimeSymbol symbol="time"/>
</Time>
<CellTypes>
<CellType class="biological" name="cells">
<Property symbol="maxCellSize" value="rand_norm(220,50)"/>
<Property symbol="minCellSize" value="max(init_volume/2,30)"/>
<Property symbol="volume" value="(maxCellSize-minCellSize)/(1+exp(-0.0005*((time-time_of_birth)-10000)))+minCellSize"/>
<Property symbol="p" name="proliferation rate" value="9e-6
"/>
<Property symbol="c" name="color" value="0"/>
<VolumeConstraint strength="1" target="volume"/>
<SurfaceConstraint strength="1" target="0.9" mode="aspherity"/>
<System solver="Euler [fixed, O(1)]" time-step="1.0">
<Rule symbol-ref="c">
<Expression>if( c > 0, c-1, 0)</Expression>
</Rule>
<Rule symbol-ref="volume">
<Expression>(maxCellSize-minCellSize)/(1+exp(-0.0005*((time-time_of_birth)-10000)))+minCellSize</Expression>
</Rule>
<Rule symbol-ref="progenitor">
<Expression>if(progenitor == 0,0,progenitor-1)</Expression>
</Rule>
<Rule symbol-ref="cc">
<Expression>if(progenitor >0, 2, c>0)</Expression>
</Rule>
</System>
<CellDivision daughterID="daughter" division-plane="random" write-log="csv">
<Condition>p>rand_uni(0,1) and((time-time_of_birth)==0 or ((time-time_of_birth)>cellCycleTime))</Condition>
<Triggers>
<Rule symbol-ref="c" name="color after division">
<Expression>1900</Expression>
</Rule>
<Rule symbol-ref="p">
<Expression>if(mother2daughters<p_rediv,p=1,p=9e-6)
</Expression>
</Rule>
<Rule symbol-ref="time_of_birth">
<Expression>time</Expression>
</Rule>
<Rule symbol-ref="cellCycleTime">
<Expression>(-(ln(rand_uni(0,1)))*85*100)+2215</Expression>
</Rule>
<!-- <Disabled>
<Rule symbol-ref="progenitor">
<Expression>if(daughter ==2 and sym_division >0.9,abs((mother2daughters<0.45)-1)*rint(cellCycleTime),(mother2daughters<0.45)*rint(cellCycleTime))
</Expression>
</Rule>
</Disabled>
-->
<Rule symbol-ref="minCellSize">
<Expression>volume/2</Expression>
</Rule>
<Rule symbol-ref="maxCellSize">
<Expression>rand_norm(220,50)</Expression>
</Rule>
<!-- <Disabled>
<Rule symbol-ref="cellCycleTime">
<Expression>if(daughter==2,rand_norm((-(ln(mother2daughters))*85*100)+2215,300),(-(ln(mother2daughters))*85*100)+2215)</Expression>
</Rule>
</Disabled>
-->
<Rule symbol-ref="progenitor">
<Expression>if(rand_uni(0,1)<0.10,rint(cellCycleTime),0)</Expression>
</Rule>
<Rule symbol-ref="mother2daughters">
<Expression>rand_uni(0,1)</Expression>
</Rule>
<Rule symbol-ref="m2dProg">
<Expression>rand_uni(0,1)</Expression>
</Rule>
</Triggers>
</CellDivision>
<CellDeath>
<Condition>progenitor ==1 or (progenitor >1 and abs((time-time_of_birth)-cellCycleTime) <3 )</Condition>
</CellDeath>
<Property symbol="clone" value="rand_uni(1,255)"/>
<Property symbol="initialId" value="cell.id"/>
<Property symbol="time_of_birth" value="0.0"/>
<Property symbol="cellCycleTime" value="(-(ln(rand_uni(0,1)))*85*100)+2215"/>
<Property symbol="progenitor" value="0.0"/>
<Property symbol="mother2daughters" value="rand_uni(0,1)"/>
<Property symbol="sym_division" value="rand_uni(0,1)"/>
<Property symbol="cc" value="1"/>
<Property symbol="m2dProg" value="rand_uni(0,1)"/>
</CellType>
</CellTypes>
<CPM>
<Interaction default="0">
<Contact type1="cells" type2="cells" value="-4"/>
</Interaction>
<MonteCarloSampler stepper="edgelist">
<MCSDuration value="1.0"/>
<Neighborhood>
<Order>2</Order>
</Neighborhood>
<MetropolisKinetics yield="0.1" temperature="3"/>
</MonteCarloSampler>
<ShapeSurface scaling="norm">
<Neighborhood>
<Order>3</Order>
</Neighborhood>
</ShapeSurface>
</CPM>
<CellPopulations>
<Population type="cells" size="1">
<InitCircle number-of-cells="500" mode="random">
<Dimensions radius="100" center="750,750,0"/>
</InitCircle>
</Population>
</CellPopulations>
<Analysis>
<Gnuplotter time-step="1000" decorate="false">
<Terminal name="pdf"/>
<Plot>
<Cells min="0.0" max="1" value="c">
<ColorMap>
<Color color="red" value="1"/>
<Color color="green" value="0.0"/>
</ColorMap>
</Cells>
<!-- <Disabled>
<CellLabels fontsize="4" value="0.0"/>
</Disabled>
-->
</Plot>
<Plot>
<Cells value="cc">
<ColorMap>
<Color color="red" value="1"/>
<Color color="green" value="0.0"/>
<Color color="black" value="2"/>
</ColorMap>
</Cells>
<!-- <Disabled>
<CellLabels precision="0" fontsize="1" value="clone"/>
</Disabled>
-->
</Plot>
</Gnuplotter>
<Logger time-step="1000">
<Input>
<Symbol symbol-ref="celltype.cells.size"/>
<Symbol symbol-ref="cell.center.x"/>
<Symbol symbol-ref="cell.center.y"/>
<Symbol symbol-ref="cellCycleTime"/>
<Symbol symbol-ref="progenitor"/>
<Symbol symbol-ref="cell.volume"/>
<Symbol symbol-ref="clone"/>
<!-- <Disabled>
<Symbol symbol-ref="minCellSize"/>
</Disabled>
-->
<!-- <Disabled>
<Symbol symbol-ref="maxCellSize"/>
</Disabled>
-->
<Symbol symbol-ref="p"/>
</Input>
<Output>
<TextOutput/>
</Output>
<Plots>
<Plot time-step="1000" log-commands="true">
<Style style="linespoints" grid="true" point-size="0.5"/>
<Terminal terminal="png"/>
<X-axis maximum="stoptime" minimum="0">
<Symbol symbol-ref="time"/>
</X-axis>
<Y-axis minimum="0">
<Symbol symbol-ref="celltype.cells.size"/>
</Y-axis>
</Plot>
</Plots>
</Logger>
<Gnuplotter time-step="1000" decorate="false">
<Terminal name="png"/>
<Plot>
<Cells value="clone"/>
<!-- <Disabled>
<CellLabels precision="0" fontsize="1" fontcolor="white" value="clone"/>
</Disabled>
-->
</Plot>
<Plot>
<Cells value="cc">
<ColorMap>
<Color color="red" value="1"/>
<Color color="green" value="0.0"/>
<Color color="black" value="2"/>
</ColorMap>
</Cells>
<!-- <Disabled>
<CellLabels precision="0" fontsize="1" value="clone"/>
</Disabled>
-->
</Plot>
</Gnuplotter>
<!-- <Disabled>
<DependencyGraph format="svg"/>
</Disabled>
-->
<ModelGraph include-tags="#untagged" format="dot" reduced="false"/>
</Analysis>
</MorpheusModel>

Downloads
Files associated with this model: