MDCK EMT A
Multiscale simulation of EMT by minimal signaling circuit
Introduction
We model epithelial-mesenchymal transition (EMT) by first assembling an ODE model for intracellular Yes-associated protein (YAP) signalling and then embedding this single cell model within individual cells in a multiscale simulation.
Description
A simple single cell ODE model describing the YAP, Rac1, and E-cadherin interaction is constructed. Building upon several assumptions, this single cell model is adapted to a multiscale simulation of a two-dimensional wound-healing experiment based on Park et al. (2019). Nanoridge arrays (NRAs) provide directional cues to epithelial sheet expansion.
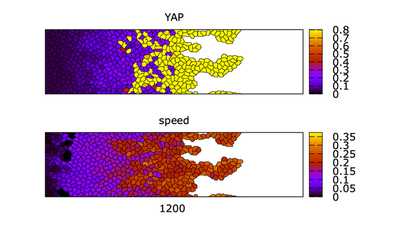
Results
Video of the model simulation YREsheetNRA_main.xml ( | ): Formation of fingers in the multiscale simulation of a control cell sheet on topographic (NRA) substrate showing sheet morphology colored for YAP (top) and cell speed (bottom) (StopTime = 1500,
The simulated cell trajactories shown below align qualitatively with the experimental results from Park et al. (2019) on substrates.
; right, **NRA** substrate: [`CellTrajCntr_NRA.xml`](#downloads).](https://d33wubrfki0l68.cloudfront.net/cffd766a3a682b9d5063bbacb56fab28a920cb5f/c0c0b/media/model/m7121/fig3_hub630a41ddb04c6e9bf6170d90fbdaf6d_1195484_137abb45efdf62bcb1469c3f50553a4e.png)
CellTrajCntr_Flat.xml; right, NRA substrate: CellTrajCntr_NRA.xml.
Video of the model simulation CellTrajCntr_Flat.xml ( | ): Trajectories for cell sheets grown on flat substrates. Speed scaled to experimentally observed range (see also Fig. 3 in the referenced paper).
Video of the model simulation CellTrajCntr_NRA.xml ( | ): Trajectories for cell sheets grown on NRA substrates. Speed scaled to experimentally observed range (see also Fig. 3 in the referenced paper).
Reference
This model is the original used in the publication, up to technical updates:
N. Mukhtar, E. N. Cytrynbaum, L. Edelstein-Keshet: A multiscale computational model of YAP signaling in epithelial fingering behavior. Biophys. J. 121 (10): 1940-1948, 2022.
Model
YREsheetNRA_main.xml
XML Preview
<?xml version='1.0' encoding='UTF-8'?>
<MorpheusModel version="4">
<Description>
<Details>Naba Mukhtar, Eric N Cytrynbaum, and Leah Edelstein-Keshet (2022) A Multiscale computational model of YAP signaling in epithelial fingering behaviour.
This xml file was used to produce Figure 2, SI Figure 6, 10
A multiscale model of a control cell sheet on nano-ridge arrays with the YAP-Rac-E cadherin circuit based on experiments by
Park, J., Kim, D.H., Shah, S.R., Kim, H.N., Kim, P., Quiñones-Hinojosa, A. and Levchenko, A., 2019. Switch-like enhancement of epithelial-mesenchymal transition by YAP through feedback regulation of WT1 and Rho-family GTPases. Nature communications, 10(1), pp.1-15.
The sheet morphology over 15 hrs and 400 micro-meters distance . On nano-ridge arrays (NRAs) a few edge cells get a high Rac1 activation rate to mimic the mechanostimulation by the topography.
This simulation is the "control" YAP case.
We thank Lutz Brusch for providing a basic cell sheet simulation that we modified and adapted to this project </Details>
<Title>YREsheetNRA</Title>
</Description>
<Space>
<Lattice class="square">
<Neighborhood>
<Order>2</Order>
</Neighborhood>
<Size value="400, 100, 0" symbol="size"/>
<BoundaryConditions>
<Condition boundary="x" type="constant"/>
<Condition boundary="-x" type="noflux"/>
<Condition boundary="y" type="periodic"/>
<Condition boundary="-y" type="periodic"/>
</BoundaryConditions>
</Lattice>
<SpaceSymbol symbol="space"/>
</Space>
<Time>
<StartTime value="0"/>
<StopTime value="1500" symbol="stoptime"/>
<TimeSymbol symbol="time"/>
<RandomSeed value="1"/>
</Time>
<Analysis>
<Gnuplotter decorate="true" time-step="50">
<Terminal name="png"/>
<Plot>
<Cells flooding="true" value="Y">
<Disabled>
<ColorMap>
<Color value="0" color="lemonchiffon"/>
<Color value="0.05" color="light-blue"/>
<Color value="0.1" color="light-red"/>
</ColorMap>
</Disabled>
</Cells>
</Plot>
<Plot title="Rac1">
<Cells flooding="true" min="0" max="3" value="R">
<ColorMap>
<Color value="0" color="blue"/>
<Color value="3" color="red"/>
<Color value="4" color="yellow"/>
</ColorMap>
</Cells>
</Plot>
<Plot>
<Cells flooding="true" min="0" max="2" value="E">
<Disabled>
<ColorMap>
<Color value="0" color="plum"/>
<Color value="4" color="blue"/>
<Color value="7" color="cyan"/>
</ColorMap>
</Disabled>
</Cells>
</Plot>
<Plot>
<Cells flooding="true" value="d.abs">
<!-- <Disabled>
<ColorMap>
<Color value="0" color="skyblue"/>
<Color value="0.1" color="violet"/>
<Color value="0.2" color="salmon"/>
</ColorMap>
</Disabled>
-->
</Cells>
</Plot>
<!-- <Disabled>
<Plot>
<Cells value="Cr" flooding="true">
<Disabled>
<ColorMap>
<Color value="0" color="red"/>
<Color value="0" color="red"/>
</ColorMap>
</Disabled>
</Cells>
</Plot>
</Disabled>
-->
</Gnuplotter>
<Logger time-step="1.0">
<Input>
<Symbol symbol-ref="cell.center.x"/>
<Symbol symbol-ref="cell.center.y"/>
</Input>
<Output>
<TextOutput/>
</Output>
<Plots>
<Plot time-step="300">
<Style point-size="0.05" style="points"/>
<Terminal terminal="png"/>
<X-axis minimum="0.0" maximum="size.x">
<Symbol symbol-ref="cell.center.x"/>
</X-axis>
<Y-axis minimum="0.0" maximum="size.y">
<Symbol symbol-ref="cell.center.y"/>
</Y-axis>
<Color-bar minimum="0.0" palette="rainbow" maximum="0.3">
<Symbol symbol-ref="d.abs"/>
</Color-bar>
</Plot>
</Plots>
</Logger>
<ModelGraph include-tags="#untagged" format="dot" reduced="false"/>
</Analysis>
<Global>
<Constant value="0.1" name="Basal rate of YAP activation" symbol="ky"/>
<Constant value="1.8" name="E-cadherin-dependent rate of YAP deactivation" symbol="kye"/>
<Constant value="2" name="Inactivation rate of YAP" symbol="Dy"/>
<Constant value="0.9" name="Initial activation rate of E-cadherin" symbol="C"/>
<Constant value="0.9" name="YAP-dependent rate of E-cadherin expression" symbol="ke"/>
<Constant value="1" name="Dissociation constant of YAP-WT1 transcriptional constant" symbol="K"/>
<Constant value="1" name="Inactivation rate of E-cadherin" symbol="De"/>
<Constant value="3" name="Hill coefficient for E-cadherin" symbol="h"/>
<Constant value="1" name="YAP-dependent rate of Rac1 expression" symbol="kr"/>
<Constant value="0.5" name="Michaelis-Menten-like constant for Rac1" symbol="Kr"/>
<Constant value="0.5" name="Degradation rate of Rac1" symbol="Dr"/>
<Constant value="6" name="Hill coefficient for Rac1" symbol="n"/>
<Constant value="1" name="Rac activation fraction" symbol="alphaR"/>
<Constant value="1.8" name="Rac1-dependent rate of YAP activation" symbol="kyr"/>
<!-- <Disabled>
<Constant value="2" symbol="A2" name="basal adhesion">
<Annotation>E-cad blocking</Annotation>
</Constant>
</Disabled>
-->
<Constant value="12" name="Max E-cadherin adhesion constant" symbol="A2">
<Annotation>Control</Annotation>
</Constant>
<Constant value="0.85" name="E-cadherin half "saturation" " symbol="A3"/>
<Constant value="0.4" name="basal migration" symbol="C1"/>
<Constant value="4" name="Max Rac1 migration constant" symbol="C2"/>
<Constant value="3" name="Rac1 half "saturation"" symbol="C3"/>
<Constant value="100" name="max time for initial leader cells to emerge" symbol="tlim"/>
<Constant value="0.2" name="fraction of neighbourhood Cr that the cell receives each time Cr spreads to it" symbol="frac"/>
<!-- <Disabled>
<Constant value="0.5" symbol="Ytot" name="Total YAP (active (Y) + inactive)">
<Annotation>KD</Annotation>
</Constant>
</Disabled>
-->
<Constant value="1.2" name="Total YAP (active (Y) + inactive)" symbol="Ytot">
<Annotation>Control</Annotation>
</Constant>
<!-- <Disabled>
<Constant value="2" symbol="Ytot" name="Total YAP (active (Y) + inactive)">
<Annotation>OE</Annotation>
</Constant>
</Disabled>
-->
<Constant value="5" name="Total Rac1 (active (R) + inactive)" symbol="Rtot"/>
<Constant value="0.02" name="parameter that determines probability of Cr spread in each time step" symbol="shareprob"/>
<Variable value="0.0" name="E-cadherin" symbol="E"/>
<Variable value="0.0" name="Basal Rac1 activation rate" symbol="Cr"/>
<!-- <Disabled>
<Variable value="0.0" symbol="x_edge2"/>
</Disabled>
-->
<!-- <Disabled>
<Mapper name="leftmost edge cell" time-step="1.0">
<Input value="(M>0)*cell.center.x*(cell.center.x>10) + (M==0)*size.x + (M>0)*size.x*(cell.center.x<=10)"/>
<Output mapping="minimum" symbol-ref="x_edge2"/>
</Mapper>
</Disabled>
-->
</Global>
<CellTypes>
<CellType name="medium" class="medium"/>
<CellType name="dividingcell" class="biological">
<VolumeConstraint strength="1" target="50"/>
<ConnectivityConstraint/>
<SurfaceConstraint mode="aspherity" strength="1" target="1"/>
<CellDivision division-plane="major">
<Condition>(rand_uni(0,1)<0.006*(exp(-time/300)+0.2)) * (cell.center.x<40)</Condition>
<Triggers/>
<Annotation>Only cells in the first 40 pixels can divide, regardless of domain size. Before, this was size.x*0.1</Annotation>
</CellDivision>
<CellDeath name="delete cells near right domain edge">
<Condition>(cell.center.x>0.99*size.x)</Condition>
</CellDeath>
<DirectedMotion direction="1, 0.0, 0.0" name="cell migration" strength="C1+C2*R/(C3+R)"/>
<Property value="0" name="YAP" symbol="Y"/>
<Property value="10" name="E-cadherin" symbol="E"/>
<Property value="0" name="Rac1" symbol="R"/>
<Property value="0.001" symbol="Cr"/>
<Property value="0.0" name="distance from right domain edge" symbol="dist"/>
<Property value="0.0" name="Sum of instantaneous speeds each time step (times 100)" symbol="avspeed"/>
<Property value="0.0" name="Average Speed * 100" symbol="truavspeed"/>
<Property value="0.0" name="Sum of instantaneous speeds over final 200 time steps (times 100)" symbol="avspeed2"/>
<Property value="0.0" name="average speed over final 200 time steps (times 100)" symbol="truavspeed2"/>
<Property value="0.0" name="Contact with medium" symbol="M"/>
<Property value="0.0" name="number of neighbour cells" symbol="neigh"/>
<Property value="0.0" name="average neighbourhood Cr" symbol="av"/>
<PropertyVector value="0.0, 0.0, 0.0" name="speed" symbol="d"/>
<NeighborhoodReporter>
<Input value="cell.type == celltype.medium.id" scaling="length"/>
<Output mapping="sum" symbol-ref="M"/>
</NeighborhoodReporter>
<NeighborhoodReporter>
<Input value="cell.type == celltype.dividingcell.id" scaling="cell"/>
<Output mapping="sum" symbol-ref="neigh"/>
</NeighborhoodReporter>
<NeighborhoodReporter>
<Input value="Cr" scaling="cell"/>
<Output mapping="average" symbol-ref="av"/>
</NeighborhoodReporter>
<MotilityReporter time-step="50">
<Velocity symbol-ref="d"/>
</MotilityReporter>
<System solver="Dormand-Prince [adaptive, O(5)]">
<DiffEqn name="Equation for YAP" symbol-ref="Y">
<Expression>(ky+kyr*R)*(Ytot-Y) - (kye*Y*E + Dy*Y)</Expression>
</DiffEqn>
<DiffEqn name="Equation for E-cadherin" symbol-ref="E">
<Expression>C - ke*Y^h/(K^h+Y^h) - De*E</Expression>
</DiffEqn>
<DiffEqn name="Equation for Rac1" symbol-ref="R">
<Expression>alphaR*(Cr + kr*Y^n/(Kr^n+Y^n))*(Rtot-R) - Dr*R</Expression>
</DiffEqn>
<Rule name="Equation for Cr spread; at each time step before tlim, cells in contact with the medium have a small chance of being endowed with high Cr; at each time step, each cell has a small chance of having its Cr be augmented by the average neighbourhood Cr (up to a max value); cells at the left edge of the domain retain low Cr" symbol-ref="Cr">
<Expression>Cr+(0.1*(M>8)*(rand_uni(0,1)<0.008)*(time<tlim)+frac*av*(rand_uni(0,1)<shareprob))*(Cr<0.1)*(cell.center.x>20)</Expression>
</Rule>
<Rule name="distance from right domain edge" symbol-ref="dist">
<Expression>size.x - cell.center.x</Expression>
</Rule>
<Rule name="Sum of instantaneous speeds each time step (times 100)" symbol-ref="avspeed">
<Expression>avspeed+d.abs*100</Expression>
</Rule>
<Rule name="average speed over the simulation (times 100)" symbol-ref="truavspeed">
<Expression>avspeed/time</Expression>
</Rule>
<Rule name="Sum of instantaneous speeds over final 100 time steps (times 100)" symbol-ref="avspeed2">
<Expression>avspeed2 + d.abs*100*(time > stoptime-201)</Expression>
</Rule>
<Rule name="average speed over 100 time steps (times 100)" symbol-ref="truavspeed2">
<Expression>avspeed2/200</Expression>
</Rule>
</System>
</CellType>
</CellTypes>
<CellPopulations>
<Population name="Initialize cell sheet at left edge of the domain" type="dividingcell" size="1">
<!-- <Disabled>
<InitRectangle mode="regular" number-of-cells="70">
<Dimensions origin="size.x/100, 0, 0" size="size.x/100, size.y, size.z"/>
</InitRectangle>
</Disabled>
-->
<InitRectangle mode="regular" number-of-cells="70">
<Dimensions origin="0.0, 0.0, 0.0" size="4.0, size.y, 0.0"/>
</InitRectangle>
</Population>
</CellPopulations>
<CPM>
<Interaction>
<Contact type2="dividingcell" value="30" type1="dividingcell">
<AddonAdhesion name="Adhesive strength" strength="5" adhesive="A2*E/(A3+E)"/>
</Contact>
<Contact type2="medium" value="12" type1="dividingcell"/>
</Interaction>
<ShapeSurface scaling="norm">
<Neighborhood>
<Order>2</Order>
</Neighborhood>
</ShapeSurface>
<MonteCarloSampler stepper="edgelist">
<MCSDuration value="1"/>
<MetropolisKinetics temperature="1"/>
<Neighborhood>
<Order>2</Order>
</Neighborhood>
</MonteCarloSampler>
</CPM>
</MorpheusModel>

YREsheetNRA_main.xml
For more information on the models discussed here,
YREsheetNRA_main.xml( | ) (produced Fig. 2),CellTrajCntr_Flat.xml( | ) (produced Fig. 3) andCellTrajCntr_NRA.xml( | ) (produced Fig. 3),
see the referenced paper.
For information on models
nra.xml( | ) (produced Fig. 4),YREcells.xml( | ) (produced SI Fig. S3),YREodes.xml( | ) (produced SI Fig. S2) andYREsheetFlat.xml( | ) (produced SI Fig. S4)
which are here attached but not discussed, please refer to the supplemental information of the referenced paper.
Downloads
Files associated with this model: