Mixed-mode Oscillations in a Rac/Rho/Paxillin Subsystem
Persistent Identifier
Use this permanent link to cite or share this Morpheus model:
Different cell migration patterns may result from different dynamic states like wave pinning (WP), mixed-mode oscillations (MMO) and relaxation oscillations (RO) of the Rho-Rac-paxillin circuit on the cell membrane.
Introduction
Different patterns of cell migration including directional, exploratory and stationary have been observed for Chinese hamster Ovary (CHO-K1) cells and are related to a mechanistic model of cell polarity (Plazen et al., 2022). This model of 4 coupled PDEs for the Rho-Rac-paxillin circuit on the cell membrane generates different cell migration patterns that correspond to different dynamic states of the PDEs like wave pinning (WP), mixed-mode oscillations (MMO) and relaxation oscillations (RO).
Description
The original model was developed in the Morpheus 2.2 series and can be downloaded as model_2.2.xml ( | ).
In addition, it has here been updated (added Plots for R, B, kb and swapped order of function declarations for P, Iks inside CellType/System to ease initialization) and can be downloaded as model.xml ( | ) to run in the latest version of Morpheus (currently the 2.3 series).
The key parameter LK can be changed in CellType/System.
Runtime in the paper is typically $8\times10^4$ time steps while the model here stops at $5\times10^3$ time steps to give a quick impression.
Results
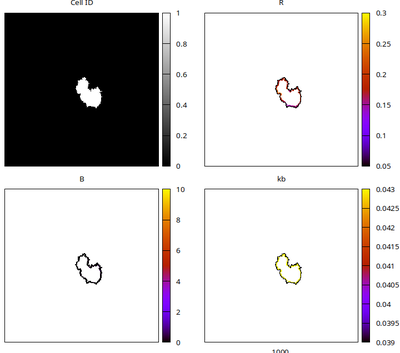
A typical simulation result is shown below.
Reference
This model is the original used in the publication, up to technical updates:
L. Plazen, J. Al Rahbani, C. M. Brown, A. Khadra: Polarity and mixed-mode oscillations may underlie different patterns of cellular migration. bioRxiv: 2022.10.31.514611, 2022.
The model originates from a preprint and thus has not been formally peer-reviewed.
Model
model.xml
XML Preview
<MorpheusModel version="4">
<Description>
<Title>Plazen2023</Title>
<Details>Full title: Mixed-mode oscillations in a Rac/Rho/Paxillin subsystem
Date: 06.01.2023
Authors: L. Plazen
ModelID: https://identifiers.org/morpheus/M7682
Software: Morpheus (open-source), download from https://morpheus.gitlab.io
Reference: This model is described in Polarity and mixed-mode oscillations may underlie different patterns of cellular migration, L. Plazen, J. Al Rahbani, C.M. Brown, A. Khadra
Currently under review
</Details>
</Description>
<Global>
<Field symbol="U" value="0"/>
<Function symbol="x">
<Expression>discr*lattice.x</Expression>
</Function>
<Function symbol="y">
<Expression>discr*lattice.y</Expression>
</Function>
</Global>
<Space>
<Lattice class="square">
<Size symbol="lattice" value="250, 250, 0"/>
<NodeLength symbol="discr" value="0.1"/>
<BoundaryConditions>
<Condition type="periodic" boundary="x"/>
<Condition type="periodic" boundary="y"/>
</BoundaryConditions>
<Neighborhood>
<Order>1</Order>
</Neighborhood>
</Lattice>
<SpaceSymbol symbol="space"/>
<MembraneLattice>
<Resolution symbol="memsize" value="200"/>
<SpaceSymbol symbol="m"/>
</MembraneLattice>
</Space>
<Time>
<StartTime value="0"/>
<StopTime value="5000"/>
<SaveInterval value="0"/>
<TimeSymbol symbol="time"/>
<RandomSeed value="19"/>
</Time>
<CellTypes>
<CellType name="cells" class="biological">
<Protrusion strength="2" field="U" maximum="1000"/>
<VolumeConstraint target="1200" strength="0.0004"/>
<SurfaceConstraint target="200" strength="0.002" mode="surface"/>
<ConnectivityConstraint/>
<MembraneProperty symbol="R" name="R" value="if((m.phi<=0.1 and m.phi>0) or (m.phi >0.3 and m.phi<0.4) or (m.phi >0.5 and m.phi<0.65) or (m.phi >0.7 and m.phi<0.72) or (m.phi >2 and m.phi<2.4) or (m.phi >3.5 and m.phi<3.7) or (m.phi >4 and m.phi<4.2) or (m.phi >5.1 and m.phi<5.7) or (m.phi >6 and m.phi<6.2) or (m.phi >7 and m.phi<7.2) , 0.3, 0.05)">
<Diffusion rate="0.0025"/>
</MembraneProperty>
<MembraneProperty symbol="RI" name="RI" value="if((m.phi<=0.1 and m.phi>0) or (m.phi >0.3 and m.phi<0.4) or (m.phi >0.5 and m.phi<0.65) or (m.phi >0.7 and m.phi<0.72) or (m.phi >2 and m.phi<2.4) or (m.phi >3.5 and m.phi<3.7) or (m.phi >4 and m.phi<4.2) or (m.phi >5.1 and m.phi<5.7) or (m.phi >6 and m.phi<6.2) or (m.phi >7 and m.phi<7.2), 0.4669, 0.8385)">
<Diffusion rate="0.43"/>
</MembraneProperty>
<MembraneProperty symbol="B" name="B" value="if((m.phi<=0.1 and m.phi>0) or (m.phi >0.3 and m.phi<0.4) or (m.phi >0.5 and m.phi<0.65) or (m.phi >0.7 and m.phi<0.72) or (m.phi >2 and m.phi<2.4) or (m.phi >3.5 and m.phi<3.7) or (m.phi >4 and m.phi<4.2) or (m.phi >5.1 and m.phi<5.7) or (m.phi >6 and m.phi<6.2) or (m.phi >7 and m.phi<7.2) , 0.5, 2)">
<Diffusion rate="0"/>
</MembraneProperty>
<MembraneProperty symbol="kb" name="kb" value="0.05">
<Diffusion rate="0"/>
</MembraneProperty>
<System time-step="0.1" name="4DPDE" solver="Euler-Maruyama [stochastic, O(1)]">
<Constant symbol="a" value="0.6281"/>
<Constant symbol="d" value="2.8773"/>
<Constant symbol="c" value="11.5"/>
<Constant symbol="Irho" name="Rho activation rate" value="0.016"/>
<Constant symbol="IR" name="Rac activation rate" value="0.0035"/>
<Constant symbol="Ik" name="Paxillin basal activation rate" value="0.009"/>
<Constant symbol="LR" name="Half max value in the Hill function of Rac" value="0.34"/>
<Constant symbol="Lrho" name="Half max value in the Hill function of Rho" value="0.34"/>
<Constant symbol="LK" name="Half max value in the Hill function of paxillin" value="5.77"/>
<Constant symbol="gamma" value="0.3"/>
<Constant symbol="epsilon" value="0.01"/>
<Constant symbol="epsilon_kb" value="0.00001"/>
<Constant symbol="gammaR" name="Parameter for the nullcline" value="8.69565"/>
<Constant symbol="alphaR" value="15"/>
<Constant symbol="alphaP" name="Linearization of Paxillin" value="2.7"/>
<Constant symbol="n" name="Non linearity level, Hill coefficient" value="4"/>
<Constant symbol="delta_P" name="Paxillin inactivation rate for Paxillin" value="0.00041"/>
<Constant symbol="delta_rho" name="Rho inactivation rate" value="0.016"/>
<Constant symbol="delta_rac" name="Rac inactivation rate" value="0.025"/>
<DiffEqn symbol-ref="R" name="PDE for active Rac">
<Expression>(IR+Iks())* (Lrho^n/ ( Lrho^n + Rho()^n) )*(RI) - delta_rac*R </Expression>
</DiffEqn>
<DiffEqn symbol-ref="RI" name="PDE for inactive Rac">
<Expression>-(IR+Iks())* (Lrho^n/ ( Lrho^n + Rho()^n) )*(RI) + delta_rac*R </Expression>
</DiffEqn>
<DiffEqn symbol-ref="B" name="PDE for B">
<Expression>dBdt</Expression>
</DiffEqn>
<DiffEqn symbol-ref="kb" name="PDE for kb">
<Expression>epsilon_kb*(0.15-R)</Expression>
</DiffEqn>
<Function symbol="dBdt" name="Derivative for B">
<Expression>epsilon*(1-kb*(B-10) -gammaR*R + 0.00001/(B+0.00001))</Expression>
</Function>
<Function symbol="K" name="Expression of K">
<Expression>alphaR*R /(1 + alphaR * R + (a*d)/(1 + d + 3/(1+1*R)))</Expression>
</Function>
<Function symbol="Rho" name="Expression of Rho in the QSSA">
<Expression>( Irho*LR^n)/( Irho*LR^n + delta_rho*( LR^n + (R + gamma*K())^n ) )</Expression>
</Function>
<Function symbol="P" name="Active Paxillin">
<Expression>B*(K()^n/(LK^n+K()^n))/( alphaP*B*(K()^n/(LK^n+K()^n)) + delta_P) </Expression>
</Function>
<Function symbol="Iks" name="Paxillin activation rate">
<Expression>Ik*(1 - 1/(1 + d + a*d*c*P + 0.5 ))</Expression>
</Function>
</System>
<Mapper>
<Input value="R-0.15"/>
<Output symbol-ref="U"/>
</Mapper>
</CellType>
</CellTypes>
<CPM>
<MonteCarloSampler stepper="edgelist">
<MCSDuration value="0.5"/>
<Neighborhood>
<Order>3</Order>
</Neighborhood>
<MetropolisKinetics yield="0.1" temperature="0.12"/>
</MonteCarloSampler>
<ShapeSurface scaling="norm">
<Neighborhood>
<Order>3</Order>
</Neighborhood>
</ShapeSurface>
<Interaction/>
</CPM>
<CellPopulations>
<Population type="cells" size="0">
<InitCellObjects mode="distance">
<Arrangement repetitions="1,1,1" displacements="0,0,0">
<Sphere center="lattice.x/2,lattice.x/2,0" radius="20"/>
</Arrangement>
</InitCellObjects>
</Population>
</CellPopulations>
<Analysis>
<Gnuplotter time-step="1000">
<Terminal name="png" size="720, 720, 0"/>
<Plot>
<Field symbol-ref="cell.id">
<ColorMap>
<Color color="white" value="1"/>
<Color color="black" value="0"/>
</ColorMap>
</Field>
</Plot>
<Plot>
<Cells max="0.3" min="0.05" value="R"/>
</Plot>
<Plot>
<Cells max="10" min="0" value="B"/>
</Plot>
<Plot>
<Cells max="0.043" min="0.039" value="kb"/>
</Plot>
</Gnuplotter>
<CellTracker time-step="5" format="ISBI 2012 (XML)"/>
<DisplacementTracker celltype="cells" time-step="1"/>
<ModelGraph include-tags="#untagged" format="dot" reduced="false"/>
</Analysis>
</MorpheusModel>

Downloads
Files associated with this model: